Bacteria Analysis
1. Pipeline: (3D) Bacteria
This pipeline segments bacteria from 3D holotomographic images and performs quantitative analysis of their morphological and optical properties. It identifies individual bacterial cells based on refractive index contrast and provides measurements such as volume, surface area, and mean RI for each object, as well as total values across the field of view.
The pipeline is suitable for analyzing bacterial populations in 3D environments, supporting applications in microbiology, infection studies, and antibiotic response assessment
Output: Bacteria mask, Bacteria instance labels, Total and individual bacteria measurements
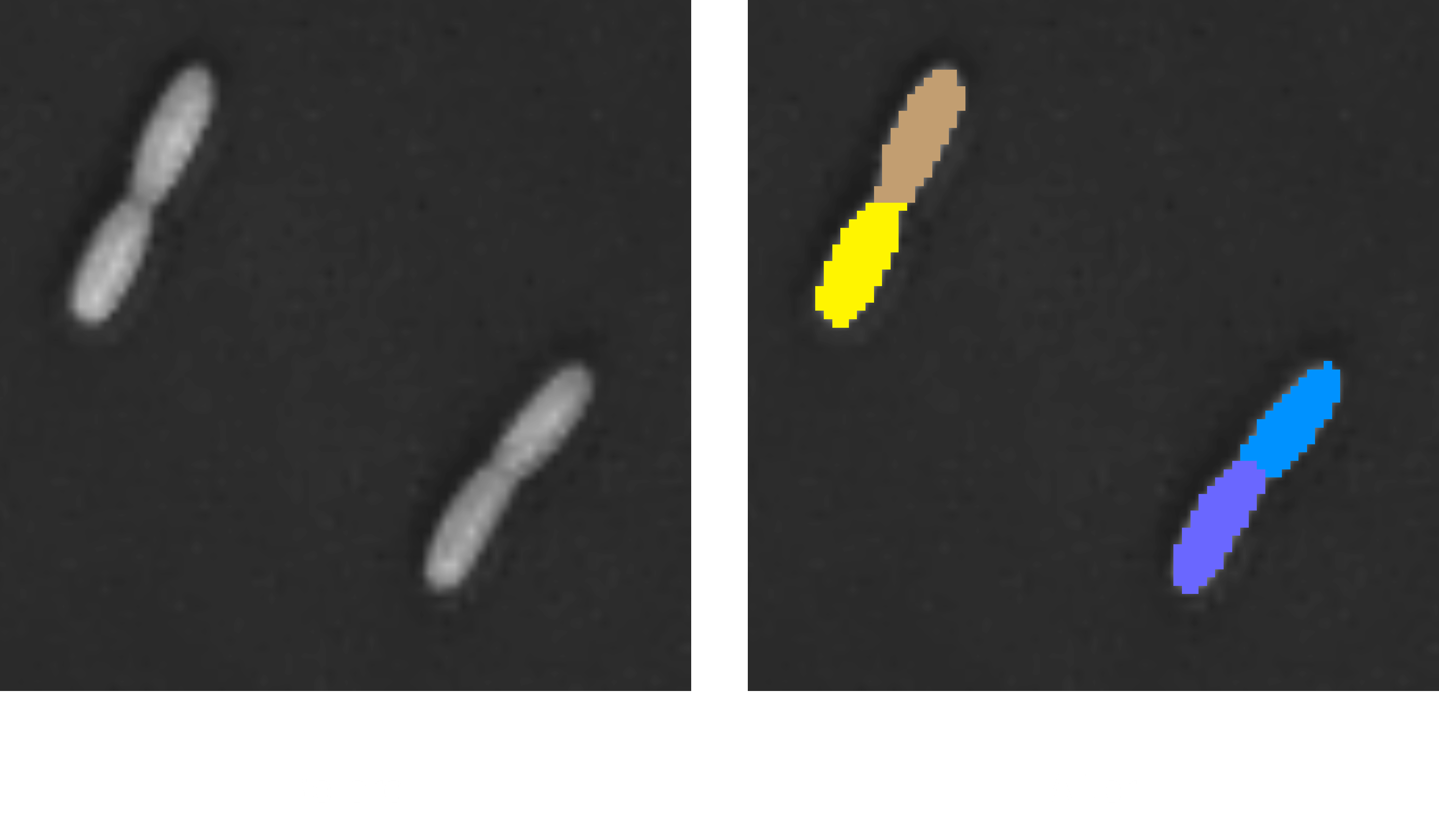
2. Pipeline: (2D) Bacteria
This pipeline segments bacteria from 2D holotomographic projections and performs quantitative analysis based on morphological and optical properties. Bacteria are identified using rule-based methods, and measurements such as area, equivalent circular diameter (ECD), and mean refractive index (RI) are provided for each instance and across the entire field of view.
It is optimized for fast and high-throughput analysis of bacterial populations in 2D, making it suitable for screening experiments and comparative studies.
Output: Bacteria mask, Bacteria instance labels, Total and individual bacteria measurements

Access to analysis pipelines is available upon request. Please send your inquiry to info@tomocube.com to explore the options best suited to your needs.