Nuclei per Cell AI-Powered
1. Pipeline: (3D) Nuclei per Cell AI-Powered
This pipeline enables per-cell analysis of nuclei in 3D by using StarDist-based segmentation for both nuclei and cells. Each nucleus is assigned to its corresponding cell based on spatial overlap, allowing accurate mapping of nuclear content within individual cytoplasmic regions.
The pipeline is designed to support single-cell analysis by providing detailed measurements of nuclei per cell. This facilitates studies on nuclear morphology, cell cycle status, and population heterogeneity.
Output: 3D masks for nuclei and cells, nucleus-to-cell assignments, per-cell nuclear measurements
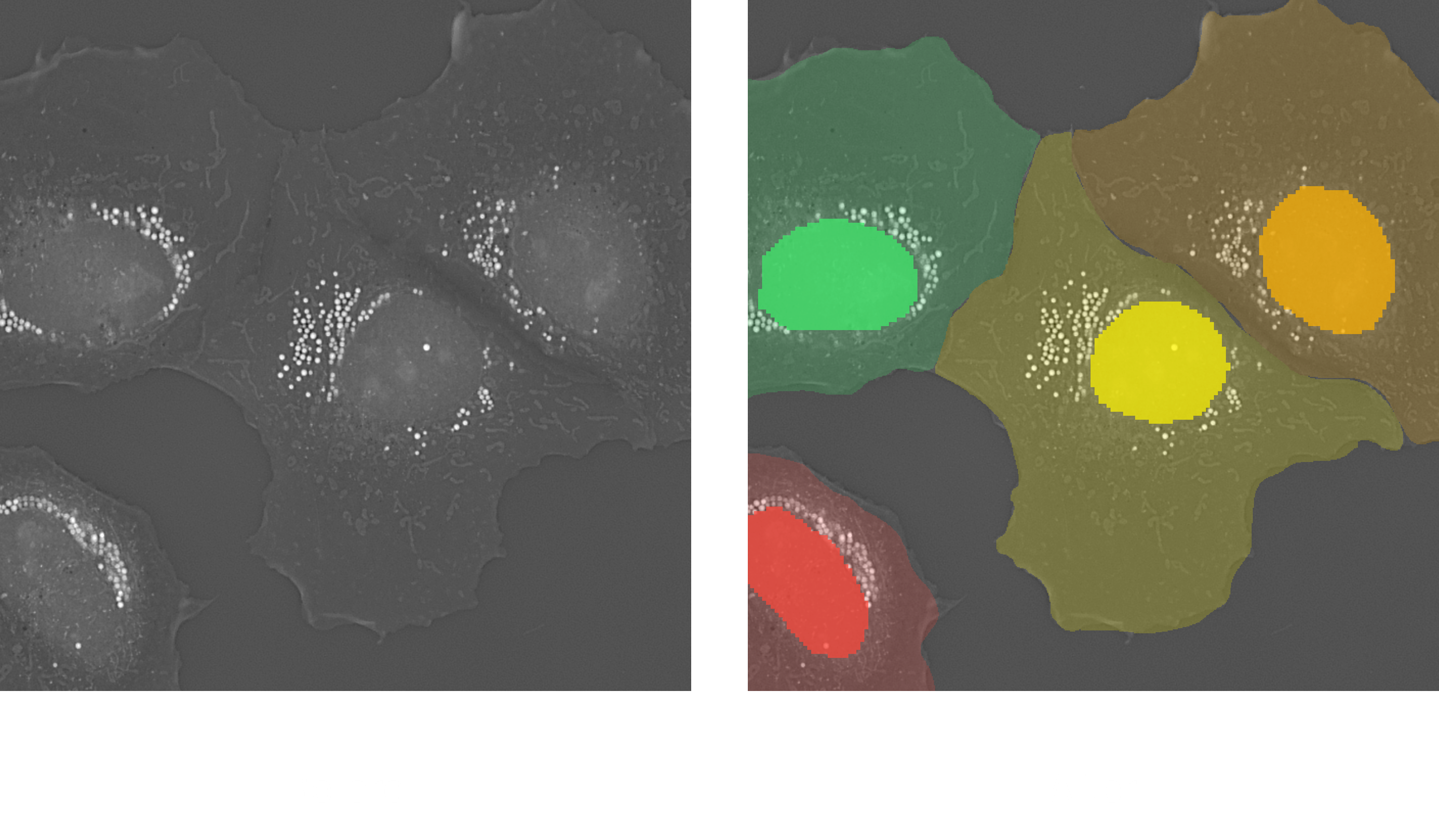
2. Pipeline: (2D) Nuclei per Cell AI-Powered
This pipeline enables per-cell analysis of nuclei in 2D using Cellpose-based segmentation. Nuclei are segmented using a Cellpose model trained to detect nuclear structures, while cell boundaries are predicted using a separate Cellpose cytoplasm model.
Each nucleus is assigned to its corresponding cell region, enabling detailed per-cell nuclear measurements such as nucleus count, area, and mean RI. This pipeline supports high-throughput single-cell analysis using holotomographic 2D projections.
Output: 2D masks for nuclei and cells, nucleus-to-cell assignments, per-cell nuclear measurements

Access to analysis pipelines is available upon request. Please send your inquiry to info@tomocube.com to explore the options best suited to your needs.